-Search query
-Search result
Showing 1 - 50 of 5,552 items for (author: ke & x)

EMDB-41441: 
CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2
Method: single particle / : Morano NC, Becker JE, Wu X, Shapiro L

PDB-8toa: 
CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2
Method: single particle / : Morano NC, Becker JE, Wu X, Shapiro L

EMDB-36659: 
Structure of human TRPV4 with antagonist A1
Method: single particle / : Fan J, Lei X

EMDB-36660: 
Structure of human TRPV4 with antagonist GSK279
Method: single particle / : Fan J, Lei X

EMDB-36675: 
Structure of human TRPV4 with antagonist A2
Method: single particle / : Fan J, Lei X

EMDB-36676: 
Structure of human TRPV4 with antagonist A2 and RhoA
Method: single particle / : Fan J, Lei X

PDB-8jvj: 
Structure of human TRPV4 with antagonist A2 and RhoA
Method: single particle / : Fan J, Lei X
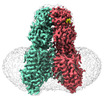
EMDB-36008: 
SIDT1 protein
Method: single particle / : Zhang JT, Jiang DH

EMDB-36009: 
transport T2
Method: single particle / : Jiang DH, Zhang JT
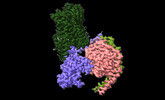
EMDB-42241: 
Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Mianserin
Method: single particle / : Zilberg G, Warren AL, Wacker D

EMDB-42245: 
Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Setiptiline
Method: single particle / : Wacker D, Parpounas AK, Warren AL, Zilberg G

PDB-8ugy: 
Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Mianserin
Method: single particle / : Zilberg G, Warren AL, Wacker D

PDB-8uh3: 
Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Setiptiline
Method: single particle / : Wacker D, Parpounas AK, Warren AL, Zilberg G

EMDB-36594: 
Cryo-EM structure of a designed AAV8-based vector
Method: single particle / : Ke X, Luo S, Zheng Q, Jiang H, Liu F, Sun X

PDB-8jre: 
Cryo-EM structure of a designed AAV8-based vector
Method: single particle / : Ke X, Luo S, Zheng Q, Jiang H, Liu F, Sun X

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-37130: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

EMDB-37131: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

PDB-8kdb: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

PDB-8kdc: 
Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form
Method: single particle / : Xie J, Wang L, Zhai G, Wu D, Lin Z, Wang M, Yan X, Gao L, Huang X, Fearns R, Chen S

EMDB-19767: 
Structure of a 2873 Scaffold Base DNA Origami V1
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19769: 
Structure of a 2873 Scaffold Base DNA Origami V2
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19770: 
Structure of a 2873 Scaffold Base DNA Origami V3
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19775: 
Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 with Desalted Purified Staples
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19776: 
Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 with HPLC Purified Staples
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19867: 
Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19874: 
Refinement Focused on the 1st Body of a 1033 Scaffold-Based DNA Origami Nanostructure V4 with TBA
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19875: 
Refinement Focused on the 2nd Body of a 1033 Scaffold-Based DNA Origami Nanostructure V4 with TBA
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-19876: 
Refinement Focused on the 3rd Body of a 1033 Scaffold-Based DNA Origami Nanostructure V4 with TBA
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

PDB-9eoq: 
Cryo-EM Structure of a 1033 Scaffold Base DNA Origami Nanostructure V4 and TBA
Method: single particle / : Ali K, Georg K, Volodymyr M, Johanna G, Maximilian NH, Lukas K, Simone C, Hendrik D

EMDB-42464: 
chEnv TTT protein in complex with 43A2 Fab
Method: single particle / : Ozorowski G, Lee WH, Ward AB

EMDB-42468: 
chEnv TTT protein in complex with CM01A Fab
Method: single particle / : Ozorowski G, Lee WH, Ward AB
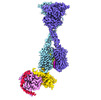
EMDB-43811: 
Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43836: 
Consensus map of human CaSR-miniGisq complex in nanodiscs
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43837: 
Local refinement map of CaSR extracellular domain in nanodisc-reconstituted human CaSR-miniGisq complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43840: 
Local refinement map of CaSR transmembrane domain in nanodisc-reconstituted human CaSR-miniGisq complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43841: 
Local refinement map of G protein in nanodisc-reconstituted human CaSR-miniGisq complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43897: 
Consensus map of human CaSR-miniGis complex in nanodiscs
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43898: 
Local refinement map of CaSR extracellular domain in nanodisc-reconstituted human CaSR-miniGis complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43899: 
Local refinement map of CaSR transmembrane domain in nanodisc-reconstituted human CaSR-miniGis complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43900: 
Local refinement map of G protein in nanodisc-reconstituted human CaSR-miniGis complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43901: 
Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43904: 
Consensus map of human CaSR-Gi3 complex in nanodiscs
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR

EMDB-43905: 
Local refinement map of CaSR extracellular domain in nanodisc-reconstituted human CaSR-Gi3 complex
Method: single particle / : Zuo H, Park J, Frangaj A, Ye J, Lu G, Manning JJ, Asher WB, Lu Z, Hu G, Wang L, Mendez J, Eng E, Zhang Z, Lin X, Grasucci R, Hendrickson WA, Clarke OB, Javitch JA, Conigrave AD, Fan QR
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search









 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
